


Reader
Publications
Group Members

JRF

JRF

JPA
Research
The theme of the lab:
The Integrative Structural Biology, Proteomics and Metabolomics Laboratory has been studying actin dynamics for nearly a decade using various biophysical methods, intending to target the actin-dependent processes.
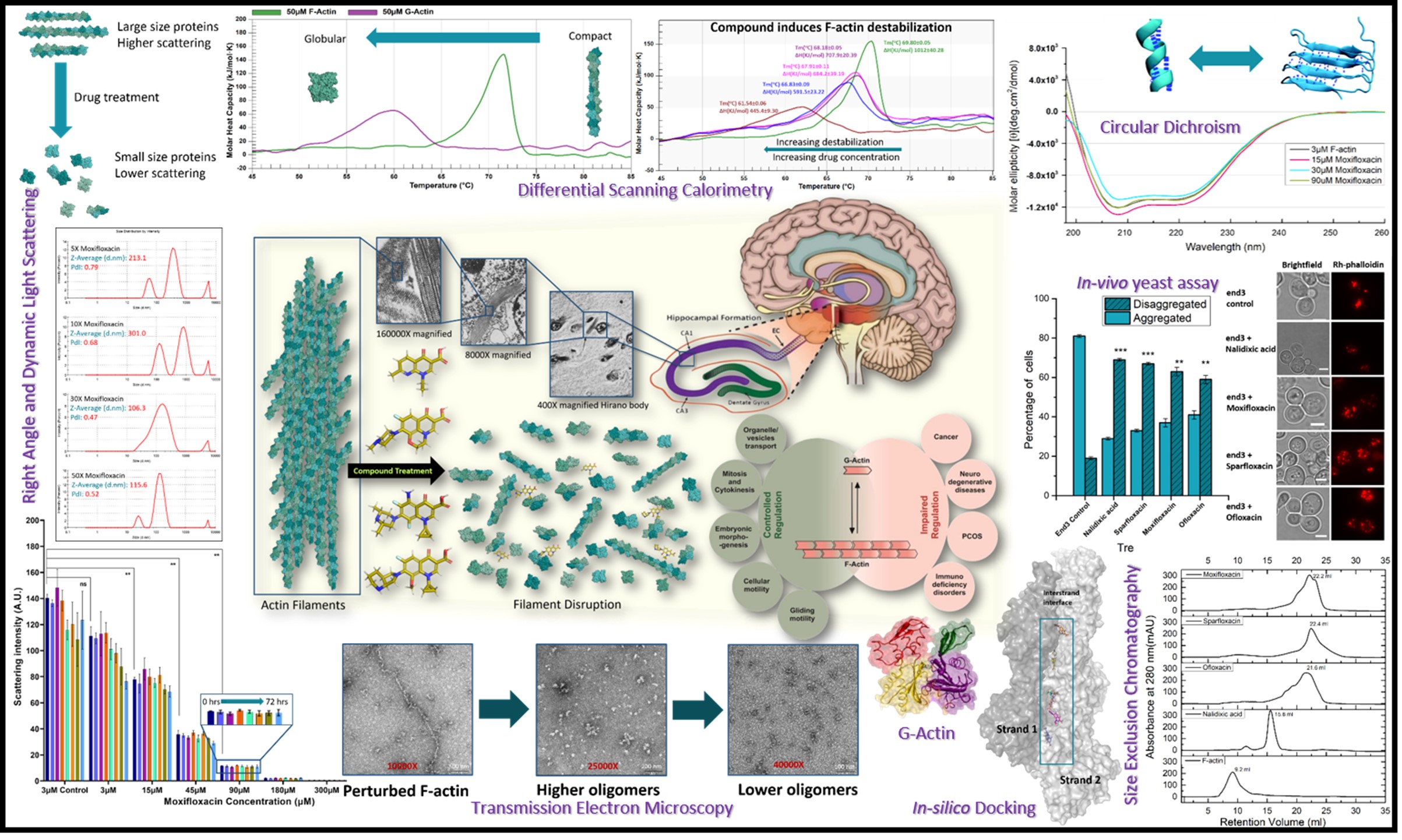
ompounds have already undergone extensive evaluation for safety and efficacy in humans. This approach significantly lowers the financial burden and reduces the risks associated with conventional drug development pipelines. Notably, our lab has previously identified alternative bioactivities of several widely used FDA-approved compounds, including members of the tetracycline family, rifampicin, colchicine, and the fluoroquinolone family, uncovering their potential as actin depolymerizers or disruptors.
Our research group is primarily a structural biology laboratory, integrating in vitro bio
Actin and actin-related proteins (ARPs) play critical roles in various human diseases due to mutations or abnormalities in their upstream and downstream signaling pathways. As a result, drug development efforts have targeted actin and ARPs. Actin-binding compounds act directly on either globular actin (G-actin) or filamentous actin (F-actin). While these actin-targeting agents have primarily been developed and tested in cancer cell lines and for the treatment of actino-pathies, none have reached clinical use due to their severe cellular toxicity.
Building on lessons from these previous challenges, our lab has focused on screening FDA-approved compounds capable of directly targeting and depolymerizing actin, with the goal of developing effective therapies for actino-pathic disorders, which currently lack viable treatment options. Drug repurposing offers a practical and cost-efficient strategy, as FDA-approved c
physical analysis and in vivo functional studies to provide a comprehensive understanding of actin dynamics at both the cellular and atomic levels. This interdisciplinary approach allows us to study actin-small molecule interactions thereby uncovering mechanistic insights that drive our drug discovery efforts.
in-vitro, in-vivo and in-silico approach to study actin-small molecule interactions
In-silico modeling, docking, and molecular dynamics (MD) simulations play a crucial role in understanding the structural dynamics and thermodynamic properties of protein-drug interactions. MD simulations of proteins and protein-ligand complexes have been instrumental in structural biology, enabling the identification of potential inhibitors based on their binding properties, as well as the structural and functional changes they induce.
Drugs from the fluoroquinolone family have been shown to exert depolymerizing effects on actin. However, key questions remain: Where does the drug bind? How does it interact with the protein? What structural or functional changes occur upon binding? And how long does the drug remain bound to its target?
MD simulations of these compounds with the pentameric actin structure have provided valuable insights into their binding dynamics, helping to identify the most and least effective inhibitors. These findings have been further validated through experimental data.
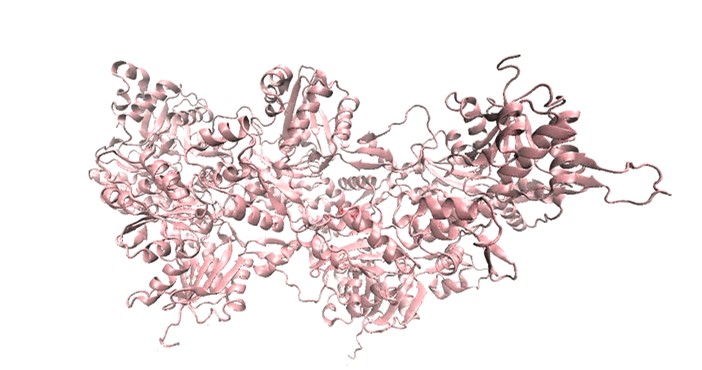
Molecular Dynamics Simulation showing the interaction between Actin and ligand
Currently, the laboratory is expanding the scope of its research by focusing on actin-regulatory proteins (ARPs). Interactions between actin and ARPs are essential for Plasmodium to execute gliding motility—a substrate-dependent mechanism employed by Plasmodium sporozoites and merozoites for host cell invasion. We aim to investigate actin-regulatory proteins to elucidate their role in actin dynamics in both in vitro and in-silico conditions. Additionally, we strive to assess whether inhibitors targeting these interactions can disrupt the function and interactions among ARPs-actin and, consequently, gliding motility.
This approach has the potential to identify small-molecule inhibitors with antimalarial activity, impairing the parasite’s ability to invade host cells and ultimately preventing clinical malaria.
The goal is to :
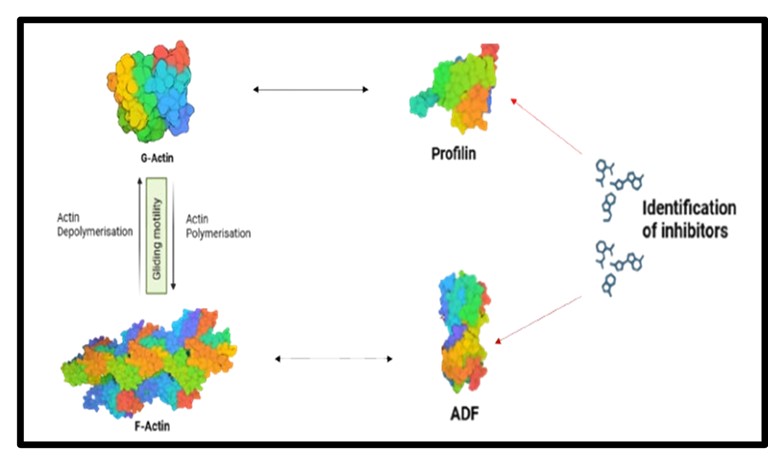
Can we inhibit the cross-talk between regulators and actin?
Our research hypothesis originates from previous mutational studies, where either depletion of ARPs in P.falciparum or P. berghei or mutation of a specific region led to complete abrogation of parasite gliding motility, impairing RBC invasion (Dans et al., "Sulfonylpiperazine Compounds," PLoS Biol, 2023; Mehta and Sibley, "Actin Depolymerizing Factor," Mol Biol Cell, 2011; Moreau et al., "Profilin-Actin Interface," PLoS Pathog, 2017)
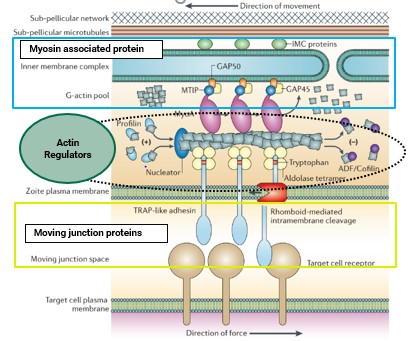
We want to identify potential inhibitors against ARPs
(Adapted from Baum et al., "Actin-Based Motility," Nat Rev Microbiol, 2006)
This study will provide the structural and biophysical basis for targeting ARPs. This investigation will address an unanswered structural understanding of ARPs and actin interactions. This would bridge the present research gap of targeting actin regulatory proteins with potential inhibitors along with high-resolution structural data. Our study involves working with ARPs from Plasmodium berghei, a model organism. The experimental approach taken to address this research questions includes techniques from the fields of molecular biology, biochemistry, biophysics and structural biology. The identified potential inhibitors will be selected for future in vivo studies. Future evaluation of their antimalarial activity in the invasion process may lead to the development of antimalarial drugs targeting gliding motility.
Thermophiles are organisms that can thrive or survive in extreme temperatures. Our focus is to identify what enables them to survive at high temperatures. We hypothesize that the synthesis of thermostable proteins and metabolites helps them adapt and proliferate in such conditions. Therefore, we are working to identify these proteins and metabolites, along with the pathways they are involved in. This will allow us to understand the fundamental mechanisms of bacterial survival in extreme environments.
The goal is to :
Below is the scheme explaining our design of experiments for metabolomics and proteomics.

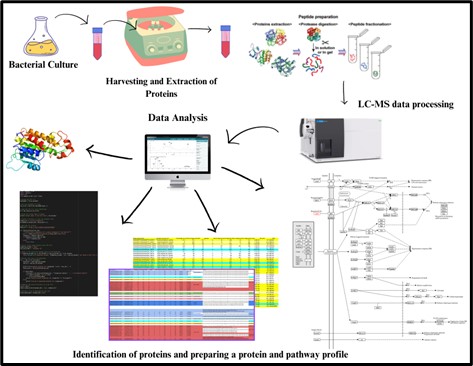
From our recent studies, we have understood that metabolic pathways exhibit interconnectedness and variability in response to differing temperature conditions. Pathway analysis revealed that sulfur metabolism and purine metabolism were ubiquitously present in all samples, regardless of inoculation methods, incubation temperatures, or bacterial life phases. Notably, the sulfur metabolism pathway triggered other carbon fixation pathways, while purine metabolism, along with other conventional pathways, was directed towards the citrate cycle. Inter-temperature pathway comparisons revealed a consistent connection between most pathways, including purine metabolism, and the citric acid cycle. A unique observation was made with Sample 3 (60_D_Log): a significant deviation from conventional pathways was noted. Except for beta-alanine metabolism, no overlap with standard pathways was observed. Instead, this sample expressed the starch and sucrose metabolism pathway, which, in turn, showed a linkage to the mycolic acid synthesis pathway. Mycolic acid is a heat-resistant lipid found in the bacterial cell envelope, and this pathway may provide a plausible explanation for the thermostability of this bacterium.
In the future, we aim to identify the proteins that play a crucial role in survival at higher temperatures.

The above poster was presented by Ms. Shruti Gupta at Aavishkar
UM-DAE Centre for Excellence in Basic Sciences
Nalanda Building, University of Mumbai, Vidyanagari, Mumbai 400098, India.
NEST/Outreach:+9186570 26481
NEST/Admissions:+9186570 26482
General Enquiries: info@cbs.ac.in
Admissions Queries:admissions@cbs.ac.in
Web: https://cbs.ac.in